
- Graph Theory - Home
- Graph Theory - Introduction
- Graph Theory - History
- Graph Theory - Fundamentals
- Graph Theory - Applications
- Types of Graphs
- Graph Theory - Types of Graphs
- Graph Theory - Simple Graphs
- Graph Theory - Multi-graphs
- Graph Theory - Directed Graphs
- Graph Theory - Weighted Graphs
- Graph Theory - Bipartite Graphs
- Graph Theory - Complete Graphs
- Graph Theory - Subgraphs
- Graph Theory - Trees
- Graph Theory - Forests
- Graph Theory - Planar Graphs
- Graph Theory - Hypergraphs
- Graph Theory - Infinite Graphs
- Graph Theory - Random Graphs
- Graph Representation
- Graph Theory - Graph Representation
- Graph Theory - Adjacency Matrix
- Graph Theory - Adjacency List
- Graph Theory - Incidence Matrix
- Graph Theory - Edge List
- Graph Theory - Compact Representation
- Graph Theory - Incidence Structure
- Graph Theory - Matrix-Tree Theorem
- Graph Properties
- Graph Theory - Basic Properties
- Graph Theory - Coverings
- Graph Theory - Matchings
- Graph Theory - Independent Sets
- Graph Theory - Traversability
- Graph Theory Connectivity
- Graph Theory - Connectivity
- Graph Theory - Vertex Connectivity
- Graph Theory - Edge Connectivity
- Graph Theory - k-Connected Graphs
- Graph Theory - 2-Vertex-Connected Graphs
- Graph Theory - 2-Edge-Connected Graphs
- Graph Theory - Strongly Connected Graphs
- Graph Theory - Weakly Connected Graphs
- Graph Theory - Connectivity in Planar Graphs
- Graph Theory - Connectivity in Dynamic Graphs
- Special Graphs
- Graph Theory - Regular Graphs
- Graph Theory - Complete Bipartite Graphs
- Graph Theory - Chordal Graphs
- Graph Theory - Line Graphs
- Graph Theory - Complement Graphs
- Graph Theory - Graph Products
- Graph Theory - Petersen Graph
- Graph Theory - Cayley Graphs
- Graph Theory - De Bruijn Graphs
- Graph Algorithms
- Graph Theory - Graph Algorithms
- Graph Theory - Breadth-First Search
- Graph Theory - Depth-First Search (DFS)
- Graph Theory - Dijkstra's Algorithm
- Graph Theory - Bellman-Ford Algorithm
- Graph Theory - Floyd-Warshall Algorithm
- Graph Theory - Johnson's Algorithm
- Graph Theory - A* Search Algorithm
- Graph Theory - Kruskal's Algorithm
- Graph Theory - Prim's Algorithm
- Graph Theory - Borůvka's Algorithm
- Graph Theory - Ford-Fulkerson Algorithm
- Graph Theory - Edmonds-Karp Algorithm
- Graph Theory - Push-Relabel Algorithm
- Graph Theory - Dinic's Algorithm
- Graph Theory - Hopcroft-Karp Algorithm
- Graph Theory - Tarjan's Algorithm
- Graph Theory - Kosaraju's Algorithm
- Graph Theory - Karger's Algorithm
- Graph Coloring
- Graph Theory - Coloring
- Graph Theory - Edge Coloring
- Graph Theory - Total Coloring
- Graph Theory - Greedy Coloring
- Graph Theory - Four Color Theorem
- Graph Theory - Coloring Bipartite Graphs
- Graph Theory - List Coloring
- Advanced Topics of Graph Theory
- Graph Theory - Chromatic Number
- Graph Theory - Chromatic Polynomial
- Graph Theory - Graph Labeling
- Graph Theory - Planarity & Kuratowski's Theorem
- Graph Theory - Planarity Testing Algorithms
- Graph Theory - Graph Embedding
- Graph Theory - Graph Minors
- Graph Theory - Isomorphism
- Spectral Graph Theory
- Graph Theory - Graph Laplacians
- Graph Theory - Cheeger's Inequality
- Graph Theory - Graph Clustering
- Graph Theory - Graph Partitioning
- Graph Theory - Tree Decomposition
- Graph Theory - Treewidth
- Graph Theory - Branchwidth
- Graph Theory - Graph Drawings
- Graph Theory - Force-Directed Methods
- Graph Theory - Layered Graph Drawing
- Graph Theory - Orthogonal Graph Drawing
- Graph Theory - Examples
- Computational Complexity of Graph
- Graph Theory - Time Complexity
- Graph Theory - Space Complexity
- Graph Theory - NP-Complete Problems
- Graph Theory - Approximation Algorithms
- Graph Theory - Parallel & Distributed Algorithms
- Graph Theory - Algorithm Optimization
- Graphs in Computer Science
- Graph Theory - Data Structures for Graphs
- Graph Theory - Graph Implementations
- Graph Theory - Graph Databases
- Graph Theory - Query Languages
- Graph Algorithms in Machine Learning
- Graph Neural Networks
- Graph Theory - Link Prediction
- Graph-Based Clustering
- Graph Theory - PageRank Algorithm
- Graph Theory - HITS Algorithm
- Graph Theory - Social Network Analysis
- Graph Theory - Centrality Measures
- Graph Theory - Community Detection
- Graph Theory - Influence Maximization
- Graph Theory - Graph Compression
- Graph Theory Real-World Applications
- Graph Theory - Network Routing
- Graph Theory - Traffic Flow
- Graph Theory - Web Crawling Data Structures
- Graph Theory - Computer Vision
- Graph Theory - Recommendation Systems
- Graph Theory - Biological Networks
- Graph Theory - Social Networks
- Graph Theory - Smart Grids
- Graph Theory - Telecommunications
- Graph Theory - Knowledge Graphs
- Graph Theory - Game Theory
- Graph Theory - Urban Planning
- Graph Theory Useful Resources
- Graph Theory - Quick Guide
- Graph Theory - Useful Resources
- Graph Theory - Discussion
Graph Theory - Link Prediction
Link Prediction
Link prediction is a task in graph theory and machine learning where the goal is to predict the existence of a link (or edge) between two nodes in a graph that is not yet present.
It assumes that relationships between nodes evolve over time and that these relationships can be predicted based on existing patterns in the graph.
Why is Link Prediction Important?
Link prediction has practical importance in various fields, such as −
- Social Networks: Suggesting new connections or friendships between users based on existing interactions.
- Recommendation Systems: Predicting which products a user may be interested in by identifying unobserved relationships.
- Bioinformatics: Predicting interactions between proteins or genes in biological networks.
- Knowledge Graphs: Adding missing relationships between entities to enhance the graph's usefulness.
Types of Link Prediction Tasks
Link prediction can be categorized based on the type of prediction being made −
- Binary Link Prediction: Predicting whether a link exists or not between two nodes.
- Top-N Link Prediction: Predicting the top N potential links that might form in the future.
Basic Concepts in Link Prediction
To understand link prediction techniques, it is important to know the fundamental graph-based concepts −
- Node Similarity: Similarity between two nodes in the graph based on their structure and attributes.
- Common Neighbors: Nodes that share common neighbors are more likely to be connected.
- Path Length: Shorter paths between nodes indicates stronger potential links.
- Graph Embeddings: Low-dimensional vector representations of graph nodes that capture structural information.
Approaches to Link Prediction
There are several approaches to link prediction, each based on different assumptions and methodologies −
Similarity-based Methods
These methods are based on the assumption that nodes that are structurally similar are likely to be connected. Common similarity measures are as follows −
- Common Neighbors: The number of common neighbors between two nodes. If two nodes have many common neighbors, they are likely to form a link.
- Jaccard Coefficient: The ratio of common neighbors to the total number of neighbors for two nodes:
Machine Learning-based Methods
These methods use graph features (e.g., node embeddings, degrees, common neighbors) as inputs to machine learning models −
- Supervised Learning: A classifier (e.g., logistic regression, random forest) is trained on a set of positive and negative links.
- Graph Embeddings: Graphs are embedded into low-dimensional spaces using techniques like DeepWalk, Node2Vec, or Graph Convolutional Networks (GCNs). These embeddings capture graph structure and node similarity for link prediction.
Probabilistic Models
Probabilistic models predict link formation by learning from graph structure and node attributes. One common approach is using matrix factorization techniques like Singular Value Decomposition (SVD) or probabilistic matrix factorization, where the goal is to predict missing values in the adjacency matrix of the graph.
Evaluation Metrics for Link Prediction
To assess the performance of link prediction algorithms, several evaluation metrics are commonly used −
- Precision: The proportion of predicted links that are correct.
- Recall: The proportion of actual links that are correctly predicted.
- F1-Score: The harmonic mean of precision and recall.
- AUC-ROC: The area under the receiver operating characteristic curve, which evaluates the ability to distinguish between positive and negative links.
Link Prediction Example Using NetworkX
In this section, we demonstrate a simple link prediction task using NetworkX and Python. We will use the common neighbors similarity method.
- Step 1: Importing Necessary Libraries
import networkx as nx from itertools import combinations
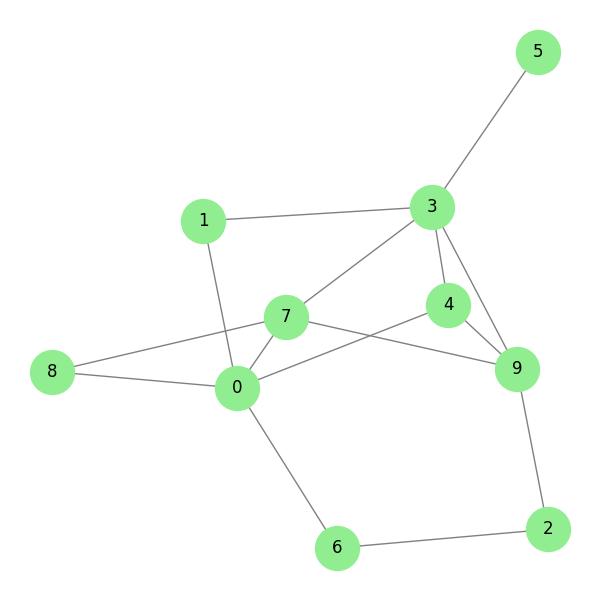
# Create a random graph with 10 nodes and 40% edge probability G = nx.erdos_renyi_graph(10, 0.4) nx.draw(G, with_labels=True)
def common_neighbors_score(G, node1, node2): common_neighbors = list(nx.common_neighbors(G, node1, node2)) return len(common_neighbors) edges = list(combinations(G.nodes, 2)) scores = [(u, v, common_neighbors_score(G, u, v)) for u, v in edges] scores_sorted = sorted(scores, key=lambda x: x[2], reverse=True) # Display top predicted links top_links = scores_sorted[:5] print(top_links)
Complete Example
Following is the complete Python code combining all steps into a single executable program −
import networkx as nx
import matplotlib.pyplot as plt
from itertools import combinations
# Step 1: Create a random graph with 10 nodes and 40% edge probability
G = nx.erdos_renyi_graph(10, 0.4)
# Draw the graph
plt.figure(figsize=(6, 6))
nx.draw(G, with_labels=True, node_color='lightblue', edge_color='gray', node_size=1000, font_size=12)
plt.title("Generated Random Graph")
plt.show()
# Step 2: Define a function to compute common neighbors similarity
def common_neighbors_score(G, node1, node2):
common_neighbors = list(nx.common_neighbors(G, node1, node2))
return len(common_neighbors)
# Step 3: Compute similarity scores for all possible node pairs
edges = list(combinations(G.nodes, 2))
scores = [(u, v, common_neighbors_score(G, u, v)) for u, v in edges]
scores_sorted = sorted(scores, key=lambda x: x[2], reverse=True)
# Step 4: Display top predicted links
top_links = scores_sorted[:5]
print("\nTop 5 Predicted Links Based on Common Neighbors:")
for link in top_links:
print(f"Nodes {link[0]} - {link[1]} have {link[2]} common neighbors")
The output will show the top 5 predicted links based on common neighbours as shown below −
Top 5 Predicted Links Based on Common Neighbors: Nodes 0 - 3 have 3 common neighbors Nodes 4 - 7 have 3 common neighbors Nodes 0 - 9 have 2 common neighbors Nodes 1 - 4 have 2 common neighbors Nodes 1 - 7 have 2 common neighbors
Link Prediction in Real-World Applications
Link prediction is used in various real-world scenarios, such as −
- Social Networks: Predicting new friendships, followers, or interactions.
- Recommendation Systems: Suggesting products, movies, or music to users based on unobserved preferences.
- Biological Networks: Predicting interactions between proteins or genes in biological systems.
- Fraud Detection: Detecting fraudulent activities in financial networks.
Challenges in Link Prediction
Despite its success, link prediction faces several challenges −
- Dynamic Graphs: The structure of the graph may evolve over time, requiring adaptive models.
- Sparse Data: Many real-world graphs are sparse, making it difficult to learn accurate models.
- Scalability: Link prediction on large-scale graphs can be computationally expensive and time-consuming.