
- Graph Theory - Home
- Graph Theory - Introduction
- Graph Theory - History
- Graph Theory - Fundamentals
- Graph Theory - Applications
- Types of Graphs
- Graph Theory - Types of Graphs
- Graph Theory - Simple Graphs
- Graph Theory - Multi-graphs
- Graph Theory - Directed Graphs
- Graph Theory - Weighted Graphs
- Graph Theory - Bipartite Graphs
- Graph Theory - Complete Graphs
- Graph Theory - Subgraphs
- Graph Theory - Trees
- Graph Theory - Forests
- Graph Theory - Planar Graphs
- Graph Theory - Hypergraphs
- Graph Theory - Infinite Graphs
- Graph Theory - Random Graphs
- Graph Representation
- Graph Theory - Graph Representation
- Graph Theory - Adjacency Matrix
- Graph Theory - Adjacency List
- Graph Theory - Incidence Matrix
- Graph Theory - Edge List
- Graph Theory - Compact Representation
- Graph Theory - Incidence Structure
- Graph Theory - Matrix-Tree Theorem
- Graph Properties
- Graph Theory - Basic Properties
- Graph Theory - Coverings
- Graph Theory - Matchings
- Graph Theory - Independent Sets
- Graph Theory - Traversability
- Graph Theory Connectivity
- Graph Theory - Connectivity
- Graph Theory - Vertex Connectivity
- Graph Theory - Edge Connectivity
- Graph Theory - k-Connected Graphs
- Graph Theory - 2-Vertex-Connected Graphs
- Graph Theory - 2-Edge-Connected Graphs
- Graph Theory - Strongly Connected Graphs
- Graph Theory - Weakly Connected Graphs
- Graph Theory - Connectivity in Planar Graphs
- Graph Theory - Connectivity in Dynamic Graphs
- Special Graphs
- Graph Theory - Regular Graphs
- Graph Theory - Complete Bipartite Graphs
- Graph Theory - Chordal Graphs
- Graph Theory - Line Graphs
- Graph Theory - Complement Graphs
- Graph Theory - Graph Products
- Graph Theory - Petersen Graph
- Graph Theory - Cayley Graphs
- Graph Theory - De Bruijn Graphs
- Graph Algorithms
- Graph Theory - Graph Algorithms
- Graph Theory - Breadth-First Search
- Graph Theory - Depth-First Search (DFS)
- Graph Theory - Dijkstra's Algorithm
- Graph Theory - Bellman-Ford Algorithm
- Graph Theory - Floyd-Warshall Algorithm
- Graph Theory - Johnson's Algorithm
- Graph Theory - A* Search Algorithm
- Graph Theory - Kruskal's Algorithm
- Graph Theory - Prim's Algorithm
- Graph Theory - Borůvka's Algorithm
- Graph Theory - Ford-Fulkerson Algorithm
- Graph Theory - Edmonds-Karp Algorithm
- Graph Theory - Push-Relabel Algorithm
- Graph Theory - Dinic's Algorithm
- Graph Theory - Hopcroft-Karp Algorithm
- Graph Theory - Tarjan's Algorithm
- Graph Theory - Kosaraju's Algorithm
- Graph Theory - Karger's Algorithm
- Graph Coloring
- Graph Theory - Coloring
- Graph Theory - Edge Coloring
- Graph Theory - Total Coloring
- Graph Theory - Greedy Coloring
- Graph Theory - Four Color Theorem
- Graph Theory - Coloring Bipartite Graphs
- Graph Theory - List Coloring
- Advanced Topics of Graph Theory
- Graph Theory - Chromatic Number
- Graph Theory - Chromatic Polynomial
- Graph Theory - Graph Labeling
- Graph Theory - Planarity & Kuratowski's Theorem
- Graph Theory - Planarity Testing Algorithms
- Graph Theory - Graph Embedding
- Graph Theory - Graph Minors
- Graph Theory - Isomorphism
- Spectral Graph Theory
- Graph Theory - Graph Laplacians
- Graph Theory - Cheeger's Inequality
- Graph Theory - Graph Clustering
- Graph Theory - Graph Partitioning
- Graph Theory - Tree Decomposition
- Graph Theory - Treewidth
- Graph Theory - Branchwidth
- Graph Theory - Graph Drawings
- Graph Theory - Force-Directed Methods
- Graph Theory - Layered Graph Drawing
- Graph Theory - Orthogonal Graph Drawing
- Graph Theory - Examples
- Computational Complexity of Graph
- Graph Theory - Time Complexity
- Graph Theory - Space Complexity
- Graph Theory - NP-Complete Problems
- Graph Theory - Approximation Algorithms
- Graph Theory - Parallel & Distributed Algorithms
- Graph Theory - Algorithm Optimization
- Graphs in Computer Science
- Graph Theory - Data Structures for Graphs
- Graph Theory - Graph Implementations
- Graph Theory - Graph Databases
- Graph Theory - Query Languages
- Graph Algorithms in Machine Learning
- Graph Neural Networks
- Graph Theory - Link Prediction
- Graph-Based Clustering
- Graph Theory - PageRank Algorithm
- Graph Theory - HITS Algorithm
- Graph Theory - Social Network Analysis
- Graph Theory - Centrality Measures
- Graph Theory - Community Detection
- Graph Theory - Influence Maximization
- Graph Theory - Graph Compression
- Graph Theory Real-World Applications
- Graph Theory - Network Routing
- Graph Theory - Traffic Flow
- Graph Theory - Web Crawling Data Structures
- Graph Theory - Computer Vision
- Graph Theory - Recommendation Systems
- Graph Theory - Biological Networks
- Graph Theory - Social Networks
- Graph Theory - Smart Grids
- Graph Theory - Telecommunications
- Graph Theory - Knowledge Graphs
- Graph Theory - Game Theory
- Graph Theory - Urban Planning
- Graph Theory Useful Resources
- Graph Theory - Quick Guide
- Graph Theory - Useful Resources
- Graph Theory - Discussion
Graph Theory - De Bruijn Graphs
De Bruijn Graphs
In graph theory, a De Bruijn graph is a directed graph that represents the relationships between substrings of a fixed length in a sequence of symbols. The graph is named after the Dutch mathematician Neels De Bruijn, who introduced it in the 1940s.
A De Bruijn graph is a directed graph where −
- The vertices represent all possible substrings of a fixed length k from a given alphabet.
- The edges represent transitions between these substrings, where each edge corresponds to appending a symbol from the alphabet to a substring.
Formally, for a given alphabet Σ and length k, the De Bruijn graph B(k, |Σ|) has −
- Vertices: Each vertex represents a distinct string of length k-1 over the alphabet Σ.
- Edges: There is a directed edge from vertex v = v1v2...vk-1 to vertex w = v2v3...vk-1a if a is a symbol from the alphabet Σ.
De Bruijn Graph Construction
To construct a De Bruijn graph, follow these steps −
- Define the alphabet Σ and the length k of the substrings.
- Identify all possible strings of length k-1 over the alphabet Σ as the vertices.
- For each vertex v = v1v2...vk-1, add a directed edge to the vertex v' = v2v3...vk-1a for each symbol a in the alphabet Σ.
For example, consider the alphabet Σ = {0, 1} and k = 3. The possible vertices of the graph are all strings of length k-1 = 2, i.e., 00, 01, 10, 11.
The corresponding De Bruijn graph B(3, 2) will have the following edges −
- 00 00 (append 0), 00 01 (append 1)
- 01 10 (append 0), 01 11 (append 1)
- 10 00 (append 0), 10 01 (append 1)
- 11 10 (append 0), 11 11 (append 1)
This process can be generalized to construct De Bruijn graphs for different alphabets and substring lengths.
Applications of De Bruijn Graphs
De Bruijn graphs are widely used in various fields, particularly in the following areas −
- Bioinformatics: In DNA sequencing, De Bruijn graphs are used to assemble genomes from short DNA fragments.
- String matching: They are used for substring search, allowing fast identification of patterns within strings.
- Data compression: De Bruijn graphs are also used in algorithms for data compression, where they help in finding repeating patterns.
- Network routing: In some network applications, De Bruijn graphs help in optimizing routing by modeling the interconnections of nodes and transitions.
Properties of De Bruijn Graphs
De Bruijn graphs have several important properties, such as −
- Symmetry: De Bruijn graphs provide a high level of symmetry due to the regularity of the vertex and edge structure.
- Degree: Every vertex in a De Bruijn graph has an in-degree and out-degree equal to the size of the alphabet, |Σ|.
- Eulerian: De Bruijn graphs are Eulerian, meaning there exists an Eulerian circuit (a closed walk that visits every edge exactly once).
Visualizing De Bruijn Graphs
Following is an example of a De Bruijn graph for the alphabet Σ = {0, 1} and length k = 3.
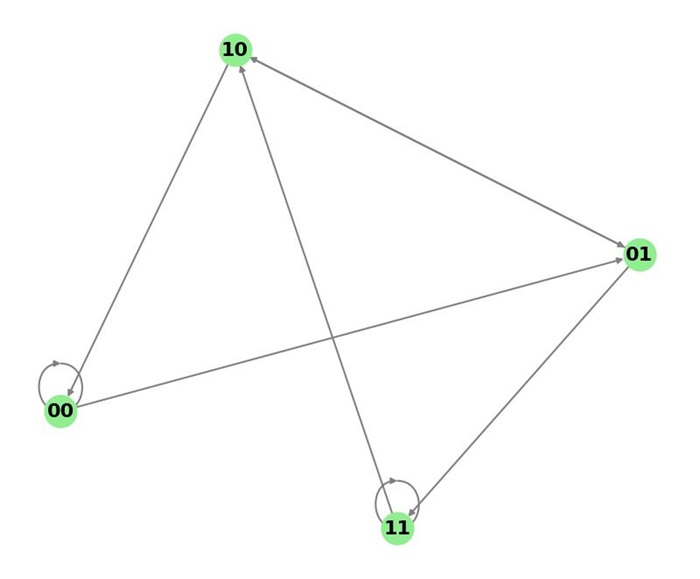
The above graph displays a De Bruijn graph for the binary alphabet {0, 1} and substrings of length 3, where the nodes represent the substrings, and the edges represent the transitions.
De Bruijn Graphs in DNA Sequencing
One of the most significant applications of De Bruijn graphs is in DNA sequencing, where they are used to assemble short DNA fragments into longer sequences. In this context −
- Each DNA fragment is represented as a vertex in the graph.
- Edges represent possible overlaps between these fragments.
- The goal is to find an Eulerian cycle in the graph that represents the original sequence of DNA.
The advantage of using De Bruijn graphs in DNA sequencing is that they provide a compact representation of the genome, reducing the complexity of the problem of assembling long DNA sequences from shorter fragments.