
 Data Structure
Data Structure Networking
Networking RDBMS
RDBMS Operating System
Operating System Java
Java MS Excel
MS Excel iOS
iOS HTML
HTML CSS
CSS Android
Android Python
Python C Programming
C Programming C++
C++ C#
C# MongoDB
MongoDB MySQL
MySQL Javascript
Javascript PHP
PHP
- Selected Reading
- UPSC IAS Exams Notes
- Developer's Best Practices
- Questions and Answers
- Effective Resume Writing
- HR Interview Questions
- Computer Glossary
- Who is Who
What is Insertional Mutagenesis
Introduction
Insertional mutagenesis is a technique used to identify and analyze genes by randomly disrupting their sequence in the genome of an organism. It is a powerful tool in molecular genetics research, as it allows researchers to identify genes responsible for various physiological processes and diseases.
In this article, we will discuss the principles and applications of insertional mutagenesis in detail.
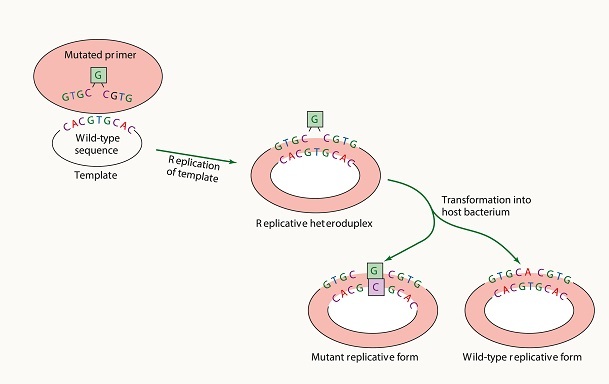
Principles of Insertional Mutagenesis
The basic principle of insertional mutagenesis involves the introduction of a foreign DNA sequence into the genome of an organism. This can be achieved by using transposons, retroviruses, or other genetic elements that can integrate into the genome. These DNA elements are modified to carry a selectable marker, such as an antibiotic resistance gene, that allows researchers to identify cells that have undergone the desired genetic alteration.
Transposons are genetic elements that can move within the genome of an organism. They can be engineered to carry a selectable marker and other genetic sequences of interest, such as promoter sequences or enhancers. Once introduced into the genome, transposons can randomly insert themselves into any location within the genome. The insertion of a transposon can disrupt a gene or alter its expression, leading to changes in the phenotype of the organism.
Retroviruses are another tool used in insertional mutagenesis. They are RNA viruses that use reverse transcriptase to convert their RNA genome into DNA, which can then integrate into the host genome. Retroviruses can be engineered to carry a selectable marker and other genetic sequences of interest, similar to transposons. Once integrated into the genome, the retroviral DNA can disrupt a gene or alter its expression, leading to changes in the phenotype of the organism.
Applications of Insertional Mutagenesis
Insertional mutagenesis has a wide range of applications in molecular genetics research. It is commonly used to identify genes involved in various physiological processes, such as development, metabolism, and disease.
Here are some of the major applications of insertional mutagenesis ?
Gene Identification
Insertional mutagenesis is a powerful tool for identifying genes responsible for various physiological processes. By randomly disrupting genes in the genome of an organism, researchers can identify genes that are essential for development, metabolism, and other biological processes. This information can be used to better understand the molecular mechanisms underlying these processes and to develop new therapies for diseases.
Disease Models
Insertional mutagenesis can be used to create animal models of human diseases. By introducing genetic mutations into the genome of an animal, researchers can create models that mimic human diseases. These models can be used to study the molecular mechanisms underlying the disease and to test potential therapies.
Cancer Research
Insertional mutagenesis has been used extensively in cancer research. Retroviruses have been used to identify genes that are involved in the development of cancer. By introducing retroviruses into the genome of an animal, researchers can identify genes that are involved in the development of tumors. This information can be used to develop new therapies for cancer.
Genetic Engineering
Insertional mutagenesis can also be used for genetic engineering. By introducing genetic sequences of interest into the genome of an organism, researchers can alter the expression of genes or create new genes. This information can be used to develop new biotechnological applications, such as the production of biofuels or pharmaceuticals.
Limitations of Insertional Mutagenesis
Although insertional mutagenesis is a powerful tool for molecular genetics research, it has some limitations. One major limitation is that it is a random process. Insertional mutagenesis can disrupt any gene in the genome, regardless of its function or importance.
This can lead to false positives, where a disrupted gene appears to be essential for a particular process when it is not.
FAQs
Q1. What is insertional mutagenesis?
Ans. Insertional mutagenesis is a technique used to identify and analyze genes by randomly disrupting their sequence in the genome of an organism. It involves the introduction of a foreign DNA sequence into the genome using transposons, retroviruses, or other genetic elements that can integrate into the genome.
Q2. What are the principles of insertional mutagenesis?
Ans. The basic principle of insertional mutagenesis involves the introduction of a foreign DNA sequence into the genome of an organism using transposons, retroviruses, or other genetic elements that can integrate into the genome. These DNA elements are modified to carry a selectable marker, such as an antibiotic resistance gene, that allows researchers to identify cells that have undergone the desired genetic alteration.
Q3. What are the applications of insertional mutagenesis?
Ans. Insertional mutagenesis has a wide range of applications in molecular genetics research. It is commonly used to identify genes involved in various physiological processes, create animal models of human diseases, study the molecular mechanisms underlying cancer, and for genetic engineering to develop new biotechnological applications.
Q4. What are the limitations of insertional mutagenesis?
Ans. One major limitation of insertional mutagenesis is that it is a random process. Insertional mutagenesis can disrupt any gene in the genome, regardless of its function or importance. This can lead to false positives, where a disrupted gene appears to be essential for a particular process when it is not.

