
 Data Structure
Data Structure Networking
Networking RDBMS
RDBMS Operating System
Operating System Java
Java MS Excel
MS Excel iOS
iOS HTML
HTML CSS
CSS Android
Android Python
Python C Programming
C Programming C++
C++ C#
C# MongoDB
MongoDB MySQL
MySQL Javascript
Javascript PHP
PHP
- Selected Reading
- UPSC IAS Exams Notes
- Developer's Best Practices
- Questions and Answers
- Effective Resume Writing
- HR Interview Questions
- Computer Glossary
- Who is Who
Parkinson Disease Prediction using Machine Learning in Python
Parkinson's Disease is a neurodegenerative disorder that affects millions worldwide, early and accurate diagnosis is crucial for effective treatment which can easily be done using machine learning in Python.
This article explores the application of machine learning techniques in predicting Parkinson's Disease using a dataset from the UCI machine learning repository. By employing the Random Forest Classifier algorithm, we demonstrate how Python can be leveraged to analyze and preprocess data, train a predictive model, and make accurate predictions.
Parkinson Disease Prediction using Machine Learning in Python
We have used Jupyter notebook to run the code in this article.
Below are the steps that we will follow for Parkinson Disease Prediction using Machine Learning in Python ?
Step 1:Import necessary libraries
Example
import pandas as pd import numpy as np import matplotlib.pyplot as plt from sklearn.metrics import accuracy_score, confusion_matrix from sklearn.preprocessing import StandardScaler from sklearn.decomposition import PCA from sklearn.model_selection import train_test_split from sklearn.ensemble import RandomForestClassifier
Step 2: Load the Parkinson's Disease dataset
The program reads the dataset from the 'parkinsons.csv' file using the pd.read_csv() function and stores it in the data variable.
Example
# Load the Parkinson's Disease dataset
data = pd.read_csv('parkinsons.csv')
Step 3: Data cleaning
The program below removes the 'name' column from the dataset using the drop() function and assigns the modified dataset back to the data variable.
Example
# Data cleaning
data = data.drop('name', axis=1) # Remove the 'name' column
Step 4: Data preprocessing
The program below separates the features (X) from the target variable (y) using the drop() function and assigns them to the respective variables.
Example
# Data preprocessing
X = data.drop('status', axis=1) # Features
y = data['status'] # Target variable
Step 5: Data analysis
The program below provides information about the dataset ?
The shape of the dataset (number of rows and columns) is printed using data.shape.
The number of samples with Parkinson's Disease and healthy samples is displayed using len(data[data['status'] == 1]) and len(data[data['status'] == 0]), respectively.
A summary of the dataset is printed using data.describe().
Example
print("Data Shape:", data.shape)
print("Parkinson's Disease Samples:", len(data[data['status'] == 1]))
print("Healthy Samples:", len(data[data['status'] == 0]))
print("\nData Summary:")
print(data.describe())
Output
Data Shape: (195, 23)
Parkinson's Disease Samples: 147
Healthy Samples: 48
Data Summary:
MDVP:Fo(Hz) MDVP:Fhi(Hz) MDVP:Flo(Hz) MDVP:Jitter(%) \
count 195.000000 195.000000 195.000000 195.000000
mean 154.228641 197.104918 116.324631 0.006220
std 41.390065 91.491548 43.521413 0.004848
min 88.333000 102.145000 65.476000 0.001680
25% 117.572000 134.862500 84.291000 0.003460
50% 148.790000 175.829000 104.315000 0.004940
75% 182.769000 224.205500 140.018500 0.007365
max 260.105000 592.030000 239.170000 0.033160
MDVP:Jitter(Abs) MDVP:RAP MDVP:PPQ Jitter:DDP MDVP:Shimmer \
count 195.000000 195.000000 195.000000 195.000000 195.000000
mean 0.000044 0.003306 0.003446 0.009920 0.029709
std 0.000035 0.002968 0.002759 0.008903 0.018857
min 0.000007 0.000680 0.000920 0.002040 0.009540
25% 0.000020 0.001660 0.001860 0.004985 0.016505
50% 0.000030 0.002500 0.002690 0.007490 0.022970
75% 0.000060 0.003835 0.003955 0.011505 0.037885
max 0.000260 0.021440 0.019580 0.064330 0.119080
max 0.685151 0.825288 -2.434031 0.450493 3.671155 0.527367
[8 rows x 23 columns]
Step 6: Data visualization
The histograms are shown using plt.show().
Example
# Data visualization data.hist(figsize=(12, 12)) plt.tight_layout() plt.show()
Output
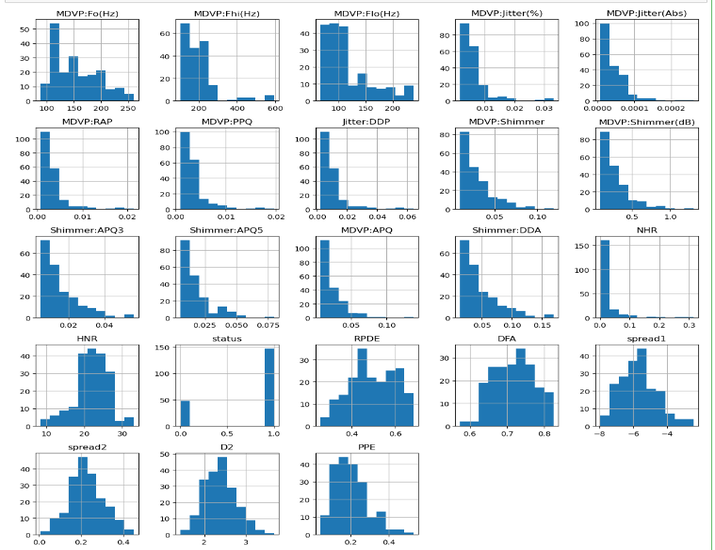
Step 7:Data scaling
The below program scales the features using StandardScaler(), which standardizes the features by subtracting the mean and scaling to unit variance. The scaled features are stored in the X_scaled variable.
Example
scaler = StandardScaler() X_scaled = scaler.fit_transform(X)
Step 8: Dimensionality reduction
It reduces the features to two principal components using PCA(n_components=2). The reduced features are stored in the X_pca variable.
Example
pca = PCA(n_components=2) X_pca = pca.fit_transform(X_scaled)
Step 9: Split the dataset into training and testing sets
The program below splits the dataset into training and testing sets using train_test_split().
Example
X_train, X_test, y_train, y_test = train_test_split(X_scaled, y, test_size=0.2, random_state=42)
Step 10: Create a Classifier known as Random Forest Classifier
The program below creates an instance of the Random Forest Classifier using RandomForestClassifier().
Train the model
Example
rf_classifier = RandomForestClassifier() # Train the model rf_classifier.fit(X_train, y_train)
Output
RandomForestClassifier()
Step 11: Make predictions on the test set
Calculate the accuracy of the model
Example
# Make predictions on the test set
y_pred = rf_classifier.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print("\nAccuracy:", accuracy)
Output
Accuracy: 0.9230769230769231
The program calculates the accuracy of the model by comparing the predicted labels (y_pred) with the actual labels (y_test).
Step 12: Confusion matrix
It uses the confusion_matrix() function from sklearn.metrics and assigns the confusion matrix to the cm variable.
Example
cm = confusion_matrix(y_test, y_pred)
print("\nConfusion Matrix:")
print(cm)
Output
Confusion Matrix: [[ 5 2] [ 1 31]]
Conclusion
In conclusion, this article presented a machine learning approach for Parkinson's Disease prediction using Python. By utilizing the Random Forest Classifier algorithm and a comprehensive dataset, we demonstrated the effectiveness of machine learning in accurately predicting the presence of Parkinson's Disease.
The results highlight the potential of this approach in assisting healthcare professionals with early diagnosis and intervention, leading to improved patient outcomes.

