
 Data Structure
Data Structure Networking
Networking RDBMS
RDBMS Operating System
Operating System Java
Java MS Excel
MS Excel iOS
iOS HTML
HTML CSS
CSS Android
Android Python
Python C Programming
C Programming C++
C++ C#
C# MongoDB
MongoDB MySQL
MySQL Javascript
Javascript PHP
PHP
- Selected Reading
- UPSC IAS Exams Notes
- Developer's Best Practices
- Questions and Answers
- Effective Resume Writing
- HR Interview Questions
- Computer Glossary
- Who is Who
Estimation of DNA Purity
Introduction
Purity of DNA refers to the DNA which is free of any protein, RNA or any other contaminant attached to it. Even after extraction of the DNA some impurities remain attached which can be removed by using certain chemical or enzymes. This step ensures the success of immediate and downstream application of DNA.
Purification of DNA
The cell is lysed by using various physical and chemical methods to release the DNA from the nucleus. Once DNA has been extracted, cellular debris remain attached to it in the form of protein, which are removed by using the proteinase enzyme or is just filtered out from the impure DNA sample.
Further purification is done by precipitating DNA using alcohols like isopropanol or ethanol. while DNA is soluble in water it cannot be separated therefore alcohols are used by stirring DNA in ethanol and spooling it out using sterile pipette. After this DNA is suspended in alkaline solution and is used.
Importance of DNA purification
Purification is important because it ensure that the same plasmid or genomic DNA is extracted which is required for the research work.
Removing the debris or contaminants from the DNA increases the shelf life of the DNA and decreases the chances of any errors during research work.
Methods of Estimating Purity of DNA
There are three methods which are generally used to estimate the purity of DNA-
Absorbance method
Fluorescence method
Gel electrophoresis
Absorbance Method Using Spectrophotometry
It is the most reliable method to test the purity and yield of the DNA. The equipment required for determination of purity by absorbance method is spectrometer which contains a UV lamp, cuvettes which are transparent to UV light and the DNA sample to be tested.
Since DNA absorbs the light strongly at a wavelength of 260nm therefore reading for absorbance is taken as 260nm or A260. This wavelength generates numbers which allow the estimation of concentration of the solution containing the DNA.
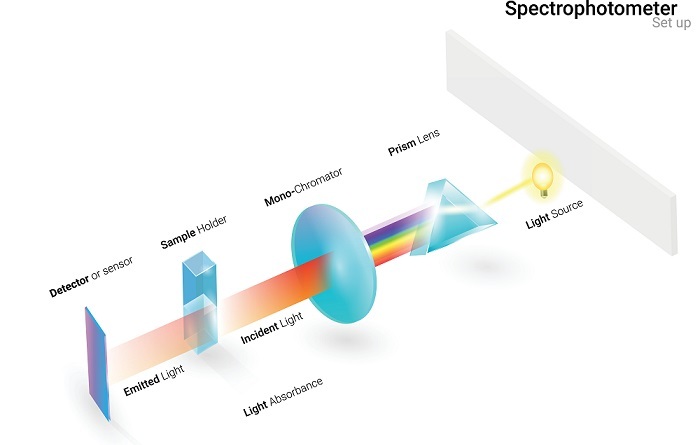
Linear range is set in the instrument to get useful numbers which is between 0.1 to 1. the turbidity of the DNA sample is measured at 320nm.
The concentration of DNA in the sample is given by the formula
Concentration of DNA(µg/ml) = (A260- A320) x dilution factor x 50µg/ml
Yield of the DNA can be measured by the multiplication of the concentration and volume of the sample.
Yield of DNA(µg) = concentration of DNA x volume of sample (ml)
But there is a limitation to this calculation not only DNA, but RNA can also absorb UV light at 260nm. Some aromatic amino acids like tyrosine, phenylalanine and tryptophan can also absorb UV light at 280nm also guanidine absorbs high at 260nm. Due to all this sometimes yield can be overestimated.
To overcome this limitation purity of the DNA is tested from 230nm to 320nm to test the presence of an impurity. Most widely used purity calculation is done by taking the ratio 260nm to 280nm. Pure DNA will have a value of 1.7 to 2.0. A value lower than 1.7 indicate the presence of other contaminants. The formula for the calculation of purity can be given as-
Purity of DNA (A260/A280) = (A260 reading- A320 reading) ÷ (A280 reading - A320 reading)
Most preferred value for pure DNA is greater than 1.5, and a value less than that indicate turbidity.
Fluorescence Method
When compared to absorbance method fluorescence method is considered more sensitive and can be used for a very small concentration of the DNA sample.
This method uses fluorescent DNA binding dyes and an instrument called fluorometer to check the purity of DNA. These dyes give more accurate results when compared to spectrophotometry.
Some of the commonly used DNA binding fluorescent dyes are Pico Green, Quantiflour and Hoechst bisbenzimidazole which specifically bind to the double stranded DNA.
The fluorometer used contains only one tube with a microplate and the samples can be read in the PCR tubes, cuvettes which makes it one of the most convenient ways to detect the purity and concentration of DNA in the sample.
Based on the dye that is chosen the excitation and emission values are set and the concentration of unknown value is calculated and compared with the known value.
The only limitation of this method is quenching and photobleaching effect of fluorescent compounds that alter the signals.
Gel Electrophoresis
Another method used to test the concentration and purity is the agarose gel electrophoresis.
This method separates the DNA based on the charge and size. Since DNA is negatively charged it will move towards the positively charged anode and smaller is the size of the DNA faster will be the movement.
The percentage at which agarose gel is prepared also decide what size rage can be separated with it. DNA bands that are formed are distinct when compared to RNA or protein which move with different rates.
The concentration can be determined by comparing the intensity of DNA band with that of standard DNA and to visualize the DNA ethidium bromide which is an intercalating dye is used.
Conclusion
When it comes to research, before the DNA sample which is isolated is used in analytical process, the quality indicators show its quality and usability. Here the quality indicators are the integrity and purity of the DNA sample. Purity largely determines the success of the research; hence the DNA sample should be free of any contaminants like proteins.

