
 Data Structure
Data Structure Networking
Networking RDBMS
RDBMS Operating System
Operating System Java
Java MS Excel
MS Excel iOS
iOS HTML
HTML CSS
CSS Android
Android Python
Python C Programming
C Programming C++
C++ C#
C# MongoDB
MongoDB MySQL
MySQL Javascript
Javascript PHP
PHP
- Selected Reading
- UPSC IAS Exams Notes
- Developer's Best Practices
- Questions and Answers
- Effective Resume Writing
- HR Interview Questions
- Computer Glossary
- Who is Who
What is Hierarchical Clustering in R Programming?
Introduction
In the vast area of data analysis and machine learning, hierarchical clustering stands as a powerful technique for grouping individuals or objects based on their similarities. When combined with the versatility and efficiency of R programming language, it becomes an even more invaluable tool for uncovering hidden patterns and structures within large datasets. In this article, we will explore what hierarchical clustering entails, dive into its various types, illustrate with a practical example, and provide a code implementation in R.
Hierarchical Clustering
Hierarchical clustering is an unsupervised learning algorithm that aims to create clusters by iteratively merging or dividing similar entities based on predetermined distance metrics. Unlike other methods like k?means clustering where we need to define the number of desired clusters beforehand, hierarchical clustering constructs a tree?like structure called a dendrogram that can be cut at a certain height to obtain multiple cluster solutions.
Types of Hierarchical Clustering
There are two main approaches when it comes to hierarchical clustering:
Agglomerative (bottom?up):This method starts by treating everyone as its own cluster and successively merges small clusters together until reaching one big cluster containing all data points. The choice of linkage criterion plays a crucial role here.
Divisive (top?down): Reverse to agglomerative approach; divisive hierarchical clustering begins with one massive cluster that contains all data points and then recursively divides them into smaller subclusters until achieving individual observations as separate clusters.
R programming to implement the Hierarchical Clustering
The Hierarchical clustering is implemented by calculating the distance with the help of the Manhattan distance.
Algorithm
Step 1:To start with, we first need to load the sample dataset.
Step 2:Before clustering, data preprocessing is necessary. We may need to standardize variables or handle missing values if present.
Step 3:Calculating distances of dissimilarity or distance between observations based on selected metrics such as Euclidean distance or Manhattan distance.
Step 4:Creating Hierarchical Clusters, that we have our distance matrix ready, we can proceed to perform hierarchical clustering using `hclust()` function in R.
Step 5:The resulting object "hc" stores all information required for subsequent steps.
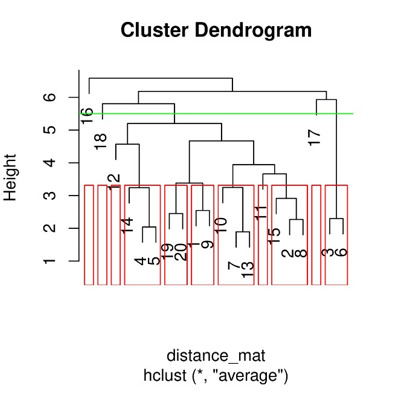
Step 6:Plotting Dendrogram, we can visualize our clusters by plotting dendrograms in R.
Example
library(cluster) # Create a sample dataset set.seed(123) x <- matrix(rnorm(100), ncol = 5) # Finding distance matrix distance_mat <- dist(x, method = 'manhattan') distance_mat # Fitting Hierarchical Clustering Model # to the training dataset set.seed(240) # Setting seed Hierar_cl <- hclust(distance_mat, method = "average") Hierar_cl # Plotting dendrogram plot(Hierar_cl) # Choosing no. of clusters # Cutting tree by height abline(h = 5.5, col = "green") # Cutting tree by no. of clusters fit <- cutree(Hierar_cl, k = 3 ) fit table(fit) rect.hclust(Hierar_cl, k = 11, border = "red")
Output
1 2 3 4 5 6 7 8
2 2.928416
3 3.820964 4.579451
4 5.870407 3.963824 6.920070
5 4.712898 3.357644 5.192501 2.041704
6 3.724940 5.188503 2.298511 7.529122 6.906090
7 4.378470 3.603915 6.073011 6.448175 6.242628 5.408591
8 2.909887 2.270199 5.993941 6.134220 5.627842 5.830570 3.531025
9 2.545686 4.500523 5.703258 5.466749 3.856739 6.130000 6.203371 4.746715
10 5.861279 5.127758 9.368977 8.324293 8.236305 8.704556 3.295965 5.013834
11 6.085281 3.179450 4.827798 5.101238 4.895692 5.436850 2.873268 4.584566
12 5.590643 2.816420 6.061696 4.307201 3.803264 6.670747 4.914120 4.986817
13 4.435100 3.563707 6.249931 6.596065 5.579229 5.771611 1.893134 3.980554
14 6.402935 4.233552 6.619402 4.029210 2.452428 8.633337 5.044223 5.761055
15 4.785512 2.544131 7.087874 5.618391 5.530404 7.696926 2.610109 3.284796
16 7.566986 7.161528 6.313095 7.075204 5.756949 6.740845 5.668959 7.578376
17 6.380095 4.860359 5.530564 7.704619 7.499072 6.327322 3.290148 5.335482
18 6.818578 4.550758 9.130209 5.378824 5.184404 9.739261 6.419430 4.339261
19 3.282054 2.655900 3.616115 5.178834 3.243561 4.331110 3.510374 3.815444
20 3.236401 2.604102 5.008448 5.395216 3.577502 6.633752 5.481153 3.662209
9 10 11 12 13 14 15 16
2
3
4
5
6
7
8
9
10 6.099033
11 6.734916 5.530202
12 6.804009 7.368849 4.130914
13 4.591857 3.200043 3.839703 4.870051
14 5.335886 6.919997 4.519932 5.595756 4.300131
15 6.393326 3.634753 3.218255 4.662948 4.028726 4.327200
16 5.414681 7.171170 5.198525 8.439225 4.010792 3.776011 7.892429
17 8.925781 6.482177 4.055843 6.170564 5.183282 6.868406 3.874253 8.586946
18 6.552868 7.159270 5.279163 5.245416 7.472245 6.473367 3.809321 9.815098
19 3.707689 6.167308 3.286156 3.779404 2.967265 4.302227 5.200031 4.681604
20 3.297399 6.324931 4.913250 4.907588 4.817754 3.963564 4.663005 6.219969
17 18 19
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18 6.804331
19 5.625103 6.543116
20 7.029360 5.821915 2.451658
Call:
hclust(d = distance_mat, method = "average")
Cluster method : average
Distance : manhattan
Number of objects: 20
[1] 1 1 2 1 1 2 1 1 1 1 1 1 1 1 1 3 2 1 1 1
fit
1 2 3
16 3 1
Conclusion
Hierarchical clustering constitutes a versatile and powerful technique for discovering underlying structures within datasets. By leveraging its practical applications and implementing it through the R programming language, we have now acquired an understanding and hands?on experience with hierarchical clustering. This approach holds immense potential for various domains like customer segmentation, image analysis, and bioinformatics assisting experts in unveiling hidden patterns that help drive informed decision?making processes.

