
 Data Structure
Data Structure Networking
Networking RDBMS
RDBMS Operating System
Operating System Java
Java MS Excel
MS Excel iOS
iOS HTML
HTML CSS
CSS Android
Android Python
Python C Programming
C Programming C++
C++ C#
C# MongoDB
MongoDB MySQL
MySQL Javascript
Javascript PHP
PHP
- Selected Reading
- UPSC IAS Exams Notes
- Developer's Best Practices
- Questions and Answers
- Effective Resume Writing
- HR Interview Questions
- Computer Glossary
- Who is Who
Reduce Data Dimensionality using PCA - Python
Any dataset used in Machine Learning algorithms may have a number of dimensions. However, not all of them contribute to giving an efficient output and simply cause the ML Model to perform poorly because of the increased size and complexity. Thus, it becomes important to eliminate such features from the dataset. To do this, we use a dimension reduction algorithm called PCA.
PCA or Principal Component Analysis helps in removing those dimensions from the dataset that do not help in optimizing the results; thereby creating a smaller and simpler dataset with most of the original and useful information. PCA is based on the concept of feature extraction, which says that when the data in a higher dimensional space is mapped to the one in lower dimensional space, the variance of the latter should be maximum.
Syntax
pca = PCA(n_components = number)
Here, PCA is the class that performs dimension reduction and pca is the object created from it. It takes only one parameter - the number of principal components we want as output.
Further, it returns the new dataset with principal components as a table when used along with the fit(), DataFrame() and head() functions as we will see in the example.
Algorithm
Step 1 ? Import Python's sklearn and pandas libraries along with the related submodules.
Step 2 ? Load the required dataset and change it to pandas dataframe.
Step 3 ? Use Standard Scaler to standardize the features and store the new dataset as a pandas data frame.
Step 4 ? Use the PCA class on the scaled dataset and create an object. Also, pass the number of components and fit and display the resultant data accordingly.
Example 1
In this example, we will take the load_diabetes dataset already present in the Python sklearn library to see how PCA is performed. In order to do this, we will use the PCA class but before that, we need to process and standardize the dataset.
#import the required libraries from sklearn import datasets #to get the load_diabetes dataset from sklearn.preprocessing import StandardScaler #this standardizes the dimensions from sklearn.decomposition import PCA #to perform PCA from sklearn.datasets import load_diabetes #the dataset that we will use to perform PCA import pandas as pd #to work with the dataframes #load the dataset as pandas dataframe diabetes = datasets.load_diabetes() df = pd.DataFrame(diabetes['data'], columns = diabetes['feature_names']) df.head() #displays the data frame when run in a different cell #standardize the dimensions by creating the object of StandardScaler scalar = StandardScaler() scaled_data = pd.DataFrame(scalar.fit_transform(df)) scaled_data #displays the dataframe after standardization when run in a different cell #apply pca pca = PCA(n_components = 4) pca.fit(scaled_data) data_pca = pca.transform(scaled_data) data_pca = pd.DataFrame(data_pca,columns=['PC1','PC2','PC3', 'PC4']) data_pca.head()
The diabetes data is loaded which returns an object similar to dictionary from where, the data is extracted into a Pandas Dataframe. We standardise the data using the StandardScaler and fit_transform() method is applied to the created Pandas Dataframe.
The fit() method is called on the standardised data, which we stored in a separate dataframe, to perform the PCA analysis. The resulting data is again stored in a separate dataframe and are printed as shown below.
Output
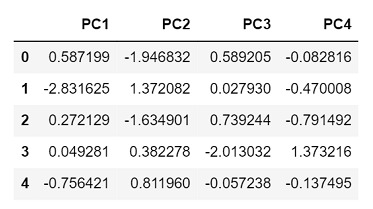
Since we chose 4 principal components, the output returned has 4 columns representing each component.
Example 2
In this example, we will take the load_wine dataset from sklearn library. Also, this time, we will set the number of principal components as only 3.
#import the required libraries from sklearn import datasets #to get the load_wine dataset from sklearn.preprocessing import StandardScaler #this standardizes the dimensions from sklearn.decomposition import PCA #to perform PCA from sklearn.datasets import load_wine #the dataset that we will use to perform PCA import pandas as pd #to work with the dataframes #load the dataset as pandas dataframe wine = datasets.load_wine() df = pd.DataFrame(wine['data'], columns = wine['feature_names']) df.head() #displays the dataframe when run in a different cell #standardize the dimensions by creating the object of StandardScaler scalar = StandardScaler() scaled_data = pd.DataFrame(scalar.fit_transform(df)) scaled_data #displays the dataframe after standardization when run in a different cell #apply pca pca = PCA(n_components = 3) pca.fit(scaled_data) data_pca = pca.transform(scaled_data) data_pca = pd.DataFrame(data_pca,columns=['PC1','PC2','PC3']) data_pca.head()
Output
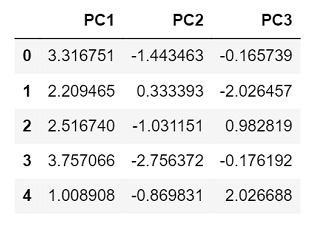
Since we chose only 3 principal components this time, the output returned has only 3 columns representing each component.
Conclusion
PCA not only makes the components of a dataset independent from each other but also solves the problem of overfitting by reducing the number of features of the dataset. However, dimension reduction is not limited to only PCA. There are other methods like LDA - Linear Discriminant Analysis and GDA - Generalized Discriminant Analysis which help in achieving the same.

