Data Structure
Data Structure Networking
Networking RDBMS
RDBMS Operating System
Operating System Java
Java MS Excel
MS Excel iOS
iOS HTML
HTML CSS
CSS Android
Android Python
Python C Programming
C Programming C++
C++ C#
C# MongoDB
MongoDB MySQL
MySQL Javascript
Javascript PHP
PHP
- Selected Reading
- UPSC IAS Exams Notes
- Developer's Best Practices
- Questions and Answers
- Effective Resume Writing
- HR Interview Questions
- Computer Glossary
- Who is Who
How to deal with warning message “Removed X rows containing missing values” for a column of an R data frame while creating a plot?
If we have missing values/NA in our data frame and create a plot using ggplot2 without excluding those missing values then we get the warning “Removed X rows containing missing values”, here X will be the number of rows for the column that contain NA values. But the plot will be correct because it will be calculated by excluding the NA’s. To avoid this error, we just need to pass the subset of the data frame column that do not contains NA values as shown in the below example.
Consider the below data frame with y column having few NA values −
Example
set.seed(112) x<-sample(0:10,25,replace=TRUE) y<-sample(c(21:25,NA),25,replace=TRUE) df<-data.frame(x,y) df
Output
x y 1 4 21 2 10 NA 3 10 23 4 10 22 5 2 NA 6 1 NA 7 0 25 8 8 NA 9 1 22 10 4 23 11 2 21 12 3 23 13 9 25 14 6 25 15 7 21 16 10 24 17 6 NA 18 6 NA 19 8 NA 20 4 24 21 1 23 22 7 21 23 1 21 24 0 22 25 4 NA
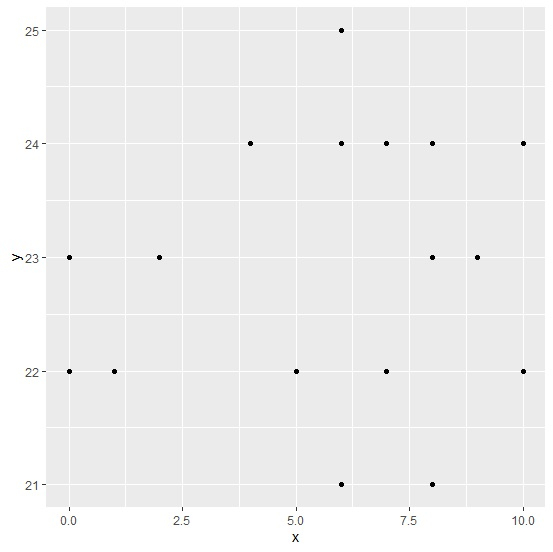
Loading ggplot2 package and creating point chart for x and y columns of df −
library(ggplot2) ggplot(df,aes(x,y))+geom_point()
Warning message −
Removed 5 rows containing missing values (geom_point) −
Here, we are getting the warning message for missing values.
Plot Output
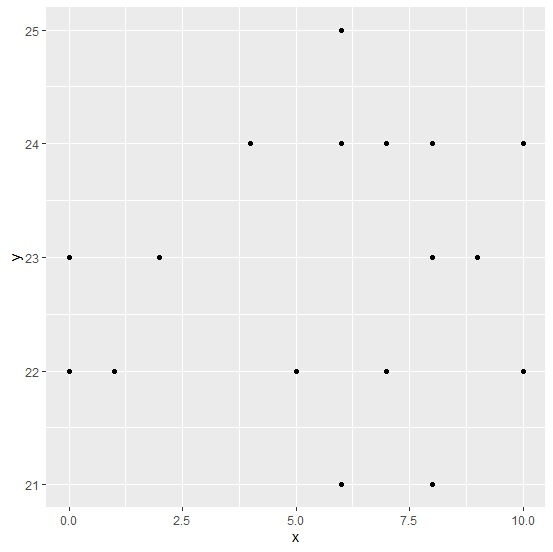
Creating the point chart for x and y by excluding the NA values −
Example
ggplot(data=subset(df,!is.na(y)),aes(x,y))+geom_point()
Output of the plot would be same as shown above but the warning message will not be there −