
- SciPy - Home
- SciPy - Introduction
- SciPy - Environment Setup
- SciPy - Basic Functionality
- SciPy - Relationship with NumPy
- SciPy Clusters
- SciPy - Clusters
- SciPy - Hierarchical Clustering
- SciPy - K-means Clustering
- SciPy - Distance Metrics
- SciPy Constants
- SciPy - Constants
- SciPy - Mathematical Constants
- SciPy - Physical Constants
- SciPy - Unit Conversion
- SciPy - Astronomical Constants
- SciPy - Fourier Transforms
- SciPy - FFTpack
- SciPy - Discrete Fourier Transform (DFT)
- SciPy - Fast Fourier Transform (FFT)
- SciPy Integration Equations
- SciPy - Integrate Module
- SciPy - Single Integration
- SciPy - Double Integration
- SciPy - Triple Integration
- SciPy - Multiple Integration
- SciPy Differential Equations
- SciPy - Differential Equations
- SciPy - Integration of Stochastic Differential Equations
- SciPy - Integration of Ordinary Differential Equations
- SciPy - Discontinuous Functions
- SciPy - Oscillatory Functions
- SciPy - Partial Differential Equations
- SciPy Interpolation
- SciPy - Interpolate
- SciPy - Linear 1-D Interpolation
- SciPy - Polynomial 1-D Interpolation
- SciPy - Spline 1-D Interpolation
- SciPy - Grid Data Multi-Dimensional Interpolation
- SciPy - RBF Multi-Dimensional Interpolation
- SciPy - Polynomial & Spline Interpolation
- SciPy Curve Fitting
- SciPy - Curve Fitting
- SciPy - Linear Curve Fitting
- SciPy - Non-Linear Curve Fitting
- SciPy - Input & Output
- SciPy - Input & Output
- SciPy - Reading & Writing Files
- SciPy - Working with Different File Formats
- SciPy - Efficient Data Storage with HDF5
- SciPy - Data Serialization
- SciPy Linear Algebra
- SciPy - Linalg
- SciPy - Matrix Creation & Basic Operations
- SciPy - Matrix LU Decomposition
- SciPy - Matrix QU Decomposition
- SciPy - Singular Value Decomposition
- SciPy - Cholesky Decomposition
- SciPy - Solving Linear Systems
- SciPy - Eigenvalues & Eigenvectors
- SciPy Image Processing
- SciPy - Ndimage
- SciPy - Reading & Writing Images
- SciPy - Image Transformation
- SciPy - Filtering & Edge Detection
- SciPy - Top Hat Filters
- SciPy - Morphological Filters
- SciPy - Low Pass Filters
- SciPy - High Pass Filters
- SciPy - Bilateral Filter
- SciPy - Median Filter
- SciPy - Non - Linear Filters in Image Processing
- SciPy - High Boost Filter
- SciPy - Laplacian Filter
- SciPy - Morphological Operations
- SciPy - Image Segmentation
- SciPy - Thresholding in Image Segmentation
- SciPy - Region-Based Segmentation
- SciPy - Connected Component Labeling
- SciPy Optimize
- SciPy - Optimize
- SciPy - Special Matrices & Functions
- SciPy - Unconstrained Optimization
- SciPy - Constrained Optimization
- SciPy - Matrix Norms
- SciPy - Sparse Matrix
- SciPy - Frobenius Norm
- SciPy - Spectral Norm
- SciPy Condition Numbers
- SciPy - Condition Numbers
- SciPy - Linear Least Squares
- SciPy - Non-Linear Least Squares
- SciPy - Finding Roots of Scalar Functions
- SciPy - Finding Roots of Multivariate Functions
- SciPy - Signal Processing
- SciPy - Signal Filtering & Smoothing
- SciPy - Short-Time Fourier Transform
- SciPy - Wavelet Transform
- SciPy - Continuous Wavelet Transform
- SciPy - Discrete Wavelet Transform
- SciPy - Wavelet Packet Transform
- SciPy - Multi-Resolution Analysis
- SciPy - Stationary Wavelet Transform
- SciPy - Statistical Functions
- SciPy - Stats
- SciPy - Descriptive Statistics
- SciPy - Continuous Probability Distributions
- SciPy - Discrete Probability Distributions
- SciPy - Statistical Tests & Inference
- SciPy - Generating Random Samples
- SciPy - Kaplan-Meier Estimator Survival Analysis
- SciPy - Cox Proportional Hazards Model Survival Analysis
- SciPy Spatial Data
- SciPy - Spatial
- SciPy - Special Functions
- SciPy - Special Package
- SciPy Advanced Topics
- SciPy - CSGraph
- SciPy - ODR
- SciPy Useful Resources
- SciPy - Reference
- SciPy - Quick Guide
- SciPy - Cheatsheet
- SciPy - Useful Resources
- SciPy - Discussion
SciPy - complete() Method
The SciPy complete() method perform the task of complete linkage(largest point) on a condensed distance matrix.
A condensed matrix is itself an array that has collection of various methods for processing the array. Also, it defines the distance between two clusters as the largest distance between any two points say(single data point in first cluster and any single data point in the second cluster).
Syntax
Following is the syntax of the SciPy complete() method −
complete(y)
Parameters
This method accepts a single parameter −
- y: This parameter store the distance of array matrix.
Return value
This method returns the linkage matrix(result).
Example 1
Following is the SciPy complete() method that shows how to perform the complete linkage clustering on a given distance matrix and visualize output using dendrogram().
import numpy as np
from scipy.cluster.hierarchy import complete, dendrogram
import matplotlib.pyplot as plt
# Distance matrix
y = np.array([0.5, 0.2, 0.3, 0.3, 0.8, 0.6])
# complete linkage clustering
result = complete(y)
# plot the dendrogram
plt.figure(figsize=(6, 4))
dendrogram(result)
plt.title('Dendrogram - Complete Linkage')
plt.xlabel('indexes')
plt.ylabel('Distance')
plt.show()
Output
The above code produces the following result −
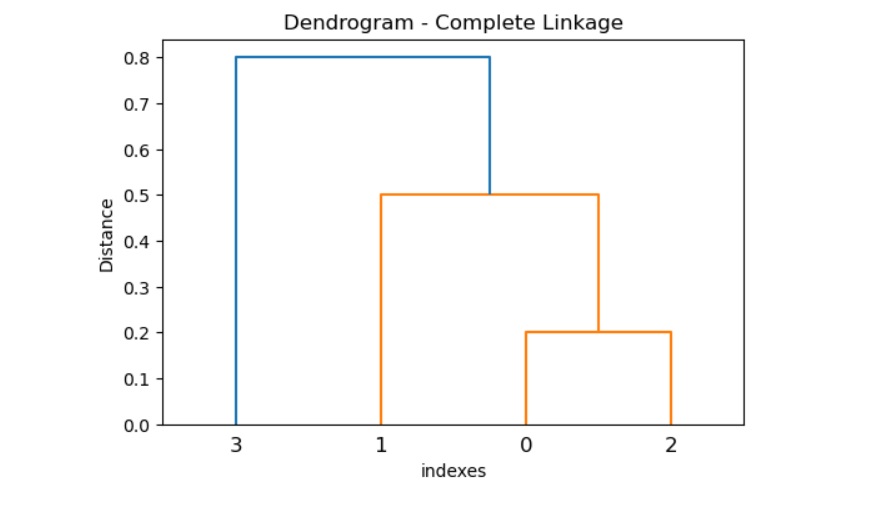
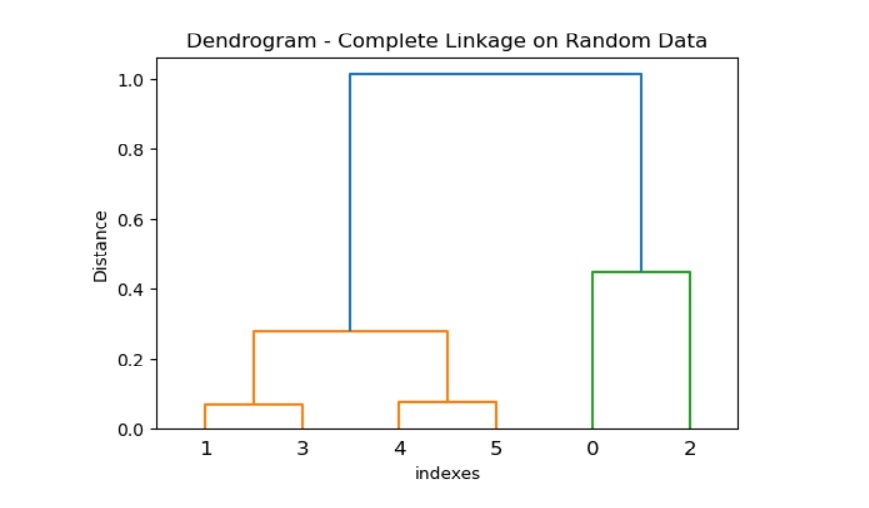
Example 2
Below the example has given a random dataset that allows the operation of a distance matrix and performs the complete linkage clustering. Then, it uses the method dendrogram () to visualize the hierarchical clustering.
import numpy as np
from scipy.spatial.distance import pdist
from scipy.cluster.hierarchy import complete, dendrogram
import matplotlib.pyplot as plt
# random data
data = np.random.rand(6, 2)
# calculate the distance matrix
y = pdist(data, metric='euclidean')
# complete linkage clustering
result = complete(y)
# plot the dendrogram
plt.figure(figsize=(6, 4))
dendrogram(result)
plt.title('Dendrogram - Complete Linkage on Random Data')
plt.xlabel('indexes')
plt.ylabel('Distance')
plt.show()
Output
The above code produces the following result −
Example 3
In this example, we illustrate the task of complete linkage clustering with the custom distance metric.
import numpy as np
from scipy.spatial.distance import pdist
from scipy.cluster.hierarchy import complete, dendrogram
import matplotlib.pyplot as plt
# Sample data
data = np.array([[1, 3], [2, 4], [8, 6], [7, 8]])
# calculate the distance matrix using a custom metric
y = pdist(data, metric='cityblock')
# complete linkage clustering
result = complete(y)
# Plot the dendrogram
plt.figure(figsize=(6, 4))
dendrogram(result)
plt.title('Dendrogram - Complete Linkage with Cityblock Distance')
plt.xlabel('Sample index')
plt.ylabel('Distance')
plt.show()
Output
The above code produces the following result −
