
- Biopython - Home
- Biopython - Introduction
- Biopython - Installation
- Creating Simple Application
- Biopython - Sequence
- Advanced Sequence Operations
- Sequence I/O Operations
- Biopython - Sequence Alignments
- Biopython - Overview of BLAST
- Biopython - Entrez Database
- Biopython - PDB Module
- Biopython - Motif Objects
- Biopython - BioSQL Module
- Biopython - Population Genetics
- Biopython - Genome Analysis
- Biopython - Phenotype Microarray
- Biopython - Plotting
- Biopython - Cluster Analysis
- Biopython - Machine Learning
- Biopython - Testing Techniques
Biopython Resources
Biopython - Motif Objects
A sequence motif is a nucleotide or amino-acid sequence pattern. Sequence motifs are formed by three-dimensional arrangement of amino acids which may not be adjacent. Biopython provides a separate module, Bio.motifs to access the functionalities of sequence motif as specified below −
from Bio import motifs
Creating Simple DNA Motif
Let us create a simple DNA motif sequence using the below command −
>>> from Bio import motifs
>>> from Bio.Seq import Seq
>>> DNA_motif = [ Seq("AGCT"),
... Seq("TCGA"),
... Seq("AACT"),
... ]
>>> seq = motifs.create(DNA_motif)
>>> print(seq)
AGCT
TCGA
AACT
To count the sequence values, use the below command −
>>> print(seq.counts)
0 1 2 3
A: 2.00 1.00 0.00 1.00
C: 0.00 1.00 2.00 0.00
G: 0.00 1.00 1.00 0.00
T: 1.00 0.00 0.00 2.00
Use the following code to count A in the sequence −
>>> seq.counts["A", :] (2.0, 1.0, 0.0, 1.0)
If you want to access the columns of counts, use the below command −
>>> seq.counts[:, 3]
{'A': 1.0, 'C': 0.0, 'G': 0.0, 'T': 2.0}
Creating a Sequence Logo
We shall now discuss how to create a Sequence Logo.
Consider the below sequence −
AGCTTACG ATCGTACC TTCCGAAT GGTACGTA AAGCTTGG
You can create your own logo using the following link − http://weblogo.berkeley.edu/
Add the above sequence and create a new logo and save the image named seq.png in your biopython folder.
seq.png
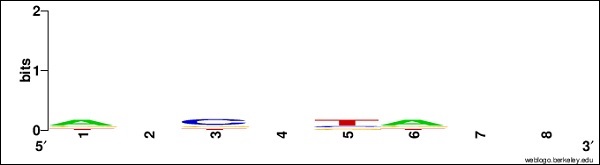
After creating the image, now run the following command −
>>> seq.weblogo("seq.png")
This DNA sequence motif is represented as a sequence logo for the LexA-binding motif.
JASPAR Database
JASPAR is one of the most popular databases. It provides facilities of any of the motif formats for reading, writing and scanning sequences. It stores meta-information for each motif. The module Bio.motifs contains a specialized class jaspar.Motif to represent meta-information attributes.
It has the following notable attributes types −
- matrix_id − Unique JASPAR motif ID
- name − The name of the motif
- tf_family − The family of motif, e.g. Helix-Loop-Helix
- data_type − the type of data used in motif.
sample.sites
Let us create a JASPAR sites format named in sample.sites in biopython folder. It is defined below −
>MA0001 ARNT 1 AACGTGatgtccta >MA0001 ARNT 2 CAGGTGggatgtac >MA0001 ARNT 3 TACGTAgctcatgc >MA0001 ARNT 4 AACGTGacagcgct >MA0001 ARNT 5 CACGTGcacgtcgt >MA0001 ARNT 6 cggcctCGCGTGc
In the above file, we have created motif instances. Now, let us create a motif object from the above instances −
>>> from Bio import motifs
>>> with open("sample.sites") as handle:
... data = motifs.read(handle,"sites")
...
>>> print(data)
TF name None
Matrix ID None
Matrix:
0 1 2 3 4 5
A: 2.00 5.00 0.00 0.00 0.00 1.00
C: 3.00 0.00 5.00 0.00 0.00 0.00
G: 0.00 1.00 1.00 6.00 0.00 5.00
T: 1.00 0.00 0.00 0.00 6.00 0.00
Here, data reads all the motif instances from sample.sites file.
Use the below command to count all the values −
>>> data.counts
{'A': [2.0, 5.0, 0.0, 0.0, 0.0, 1.0], 'C': [3.0, 0.0, 5.0, 0.0, 0.0, 0.0], 'G': [0.0, 1.0, 1.0, 6.0, 0.0, 5.0], 'T': [1.0, 0.0, 0.0, 0.0, 6.0, 0.0]}
>>>