
- Biopython - Home
- Biopython - Introduction
- Biopython - Installation
- Creating Simple Application
- Biopython - Sequence
- Advanced Sequence Operations
- Sequence I/O Operations
- Biopython - Sequence Alignments
- Biopython - Overview of BLAST
- Biopython - Entrez Database
- Biopython - PDB Module
- Biopython - Motif Objects
- Biopython - BioSQL Module
- Biopython - Population Genetics
- Biopython - Genome Analysis
- Biopython - Phenotype Microarray
- Biopython - Plotting
- Biopython - Cluster Analysis
- Biopython - Machine Learning
- Biopython - Testing Techniques
Biopython Resources
Biopython - Genome Analysis
A genome is complete set of DNA, including all of its genes. Genome analysis refers to the study of individual genes and their roles in inheritance.
Genome Diagram
Genome diagram represents the genetic information as charts. Biopython uses Bio.Graphics.GenomeDiagram module to represent GenomeDiagram. The GenomeDiagram module requires ReportLab to be installed.
Steps for creating a diagram
The process of creating a diagram generally follows the below simple pattern −
Create a FeatureSet for each separate set of features you want to display, and add Bio.SeqFeature objects to them.
Create a GraphSet for each graph you want to display, and add graph data to them.
Create a Track for each track you want on the diagram, and add GraphSets and FeatureSets to the tracks you require.
Create a Diagram, and add the Tracks to it.
Tell the Diagram to draw the image.
Write the image to a file.
example.gbk
Let us take an example of input GenBank file −
LOCUS Z78533 740 bp DNA linear PLN 30-NOV-2006
DEFINITION C.irapeanum 5.8S rRNA gene and ITS1 and ITS2 DNA.
ACCESSION Z78533
VERSION Z78533.1 GI:2765658
KEYWORDS 5.8S ribosomal RNA; 5.8S rRNA gene; internal transcribed spacer;
ITS1; ITS2.
SOURCE Cypripedium irapeanum
ORGANISM Cypripedium irapeanum
Eukaryota; Viridiplantae; Streptophyta; Embryophyta; Tracheophyta;
Spermatophyta; Magnoliophyta; Liliopsida; Asparagales; Orchidaceae;
Cypripedioideae; Cypripedium.
REFERENCE 1
AUTHORS Cox,A.V., Pridgeon,A.M., Albert,V.A. and Chase,M.W.
TITLE Phylogenetics of the slipper orchids (Cypripedioideae:
Orchidaceae): nuclear rDNA ITS sequences
JOURNAL Unpublished
REFERENCE 2 (bases 1 to 740)
AUTHORS Cox,A.V.
TITLE Direct Submission
JOURNAL Submitted (19-AUG-1996) Cox A.V., Royal Botanic Gardens, Kew,
Richmond, Surrey TW9 3AB, UK
FEATURES Location/Qualifiers
source 1..740
/organism="Cypripedium irapeanum"
/mol_type="genomic DNA"
/db_xref="taxon:49711"
misc_feature 1..380
/note="internal transcribed spacer 1"
gene 381..550
/gene="5.8S rRNA"
rRNA 381..550
/gene="5.8S rRNA"
/product="5.8S ribosomal RNA"
misc_feature 551..740
/note="internal transcribed spacer 2"
ORIGIN
1 cgtaacaagg tttccgtagg tgaacctgcg gaaggatcat tgatgagacc gtggaataaa
61 cgatcgagtg aatccggagg accggtgtac tcagctcacc gggggcattg ctcccgtggt
121 gaccctgatt tgttgttggg ccgcctcggg agcgtccatg gcgggtttga acctctagcc
181 cggcgcagtt tgggcgccaa gccatatgaa agcatcaccg gcgaatggca ttgtcttccc
241 caaaacccgg agcggcggcg tgctgtcgcg tgcccaatga attttgatga ctctcgcaaa
301 cgggaatctt ggctctttgc atcggatgga aggacgcagc gaaatgcgat aagtggtgtg
361 aattgcaaga tcccgtgaac catcgagtct tttgaacgca agttgcgccc gaggccatca
421 ggctaagggc acgcctgctt gggcgtcgcg cttcgtctct ctcctgccaa tgcttgcccg
481 gcatacagcc aggccggcgt ggtgcggatg tgaaagattg gccccttgtg cctaggtgcg
541 gcgggtccaa gagctggtgt tttgatggcc cggaacccgg caagaggtgg acggatgctg
601 gcagcagctg ccgtgcgaat cccccatgtt gtcgtgcttg tcggacaggc aggagaaccc
661 ttccgaaccc caatggaggg cggttgaccg ccattcggat gtgaccccag gtcaggcggg
721 ggcacccgct gagtttacgc
//
Install reportLab using pip if not installed.
(myenv) D:\biopython\myenv>pip3 install reportlab Collecting reportlab Obtaining dependency information for reportlab from https://files.pythonhosted.org/packages/0e/ee/5f7a31ab05cf817e0cc70ae6df51a1a4fda188c899790a3131a24dd78d18/reportlab-4.4.6-py3-none-any.whl.metadata Downloading reportlab-4.4.6-py3-none-any.whl.metadata (1.7 kB) Collecting pillow>=9.0.0 (from reportlab) Obtaining dependency information for pillow>=9.0.0 from https://files.pythonhosted.org/packages/a2/0b/d87733741526541c909bbf159e338dcace4f982daac6e5a8d6be225ca32d/pillow-12.0.0-cp312-cp312-win_amd64.whl.metadata Downloading pillow-12.0.0-cp312-cp312-win_amd64.whl.metadata (9.0 kB) Collecting charset-normalizer (from reportlab) Obtaining dependency information for charset-normalizer from https://files.pythonhosted.org/packages/3d/2d/1e5ed9dd3b3803994c155cd9aacb60c82c331bad84daf75bcb9c91b3295e/charset_normalizer-3.4.4-cp312-cp312-win_amd64.whl.metadata Using cached charset_normalizer-3.4.4-cp312-cp312-win_amd64.whl.metadata (38 kB) Downloading reportlab-4.4.6-py3-none-any.whl (2.0 MB) ââââââââââââââââââââââââââââââââââââââââ 2.0/2.0 MB 6.9 MB/s eta 0:00:00 Downloading pillow-12.0.0-cp312-cp312-win_amd64.whl (7.0 MB) ââââââââââââââââââââââââââââââââââââââââ 7.0/7.0 MB 5.3 MB/s eta 0:00:00 Using cached charset_normalizer-3.4.4-cp312-cp312-win_amd64.whl (107 kB) Installing collected packages: pillow, charset-normalizer, reportlab Successfully installed charset-normalizer-3.4.4 pillow-12.0.0 reportlab-4.4.6
We shall import all the modules first as shown below −
>>> from reportlab.lib import colors >>> from reportlab.lib.units import cm >>> from Bio.Graphics import GenomeDiagram
Now, import SeqIO module to read data −
>>> from Bio import SeqIO
record = SeqIO.read("example.gbk", "genbank")
Here, the record reads the sequence from genbank file.
Now, create an empty diagram to add track and feature set −
>>> diagram = GenomeDiagram.Diagram( "Yersinia pestis biovar Microtus plasmid pPCP1") >>> track = diagram.new_track(1, name="Annotated Features") >>> feature = track.new_set()
Let us draw a diagram for the above input records −
>>> diagram.draw(
format = "linear", orientation = "landscape", pagesize = 'A4',
... fragments = 4, start = 0, end = len(record))
>>> diagram.write("orchid.pdf", "PDF")
>>> diagram.write("orchid.eps", "EPS")
>>> diagram.write("orchid.svg", "SVG")
After executing the above command, you could see the following image saved in your Biopython directory.
genome.svg
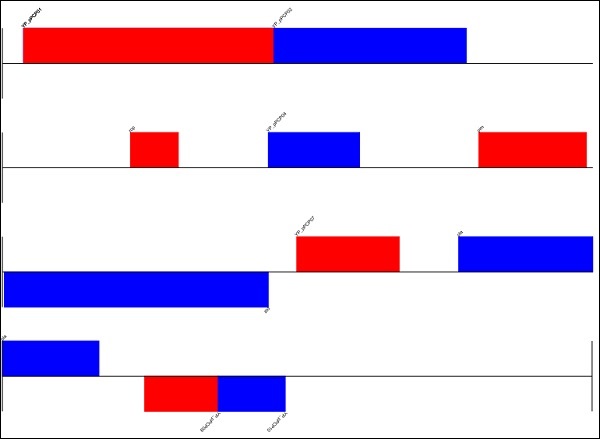
You can also draw the image in circular format by making the below changes −
>>> diagram.draw(
format = "circular", circular = True, pagesize = (20*cm,20*cm),
... start = 0, end = len(record), circle_core = 0.7)
>>> diagram.write("circular.pdf", "PDF")
Chromosomes Overview
DNA molecule is packaged into thread-like structures called chromosomes. Each chromosome is made up of DNA tightly coiled many times around proteins called histones that support its structure.
Chromosomes are not visible in the cells nucleus not even under a microscope when the cell is not dividing. However, the DNA that makes up chromosomes becomes more tightly packed during cell division and is then visible under a microscope.
In humans, each cell normally contains 23 pairs of chromosomes, for a total of 46. Twenty-two of these pairs, called autosomes, look the same in both males and females. The 23rd pair, the sex chromosomes, differ between males and females. Females have two copies of the X chromosome, while males have one X and one Y chromosome.