
- Data Structures & Algorithms
- DSA - Home
- DSA - Overview
- DSA - Environment Setup
- DSA - Algorithms Basics
- DSA - Asymptotic Analysis
- Data Structures
- DSA - Data Structure Basics
- DSA - Data Structures and Types
- DSA - Array Data Structure
- Linked Lists
- DSA - Linked List Data Structure
- DSA - Doubly Linked List Data Structure
- DSA - Circular Linked List Data Structure
- Stack & Queue
- DSA - Stack Data Structure
- DSA - Expression Parsing
- DSA - Queue Data Structure
- Searching Algorithms
- DSA - Searching Algorithms
- DSA - Linear Search Algorithm
- DSA - Binary Search Algorithm
- DSA - Interpolation Search
- DSA - Jump Search Algorithm
- DSA - Exponential Search
- DSA - Fibonacci Search
- DSA - Sublist Search
- DSA - Hash Table
- Sorting Algorithms
- DSA - Sorting Algorithms
- DSA - Bubble Sort Algorithm
- DSA - Insertion Sort Algorithm
- DSA - Selection Sort Algorithm
- DSA - Merge Sort Algorithm
- DSA - Shell Sort Algorithm
- DSA - Heap Sort
- DSA - Bucket Sort Algorithm
- DSA - Counting Sort Algorithm
- DSA - Radix Sort Algorithm
- DSA - Quick Sort Algorithm
- Graph Data Structure
- DSA - Graph Data Structure
- DSA - Depth First Traversal
- DSA - Breadth First Traversal
- DSA - Spanning Tree
- Tree Data Structure
- DSA - Tree Data Structure
- DSA - Tree Traversal
- DSA - Binary Search Tree
- DSA - AVL Tree
- DSA - Red Black Trees
- DSA - B Trees
- DSA - B+ Trees
- DSA - Splay Trees
- DSA - Tries
- DSA - Heap Data Structure
- Recursion
- DSA - Recursion Algorithms
- DSA - Tower of Hanoi Using Recursion
- DSA - Fibonacci Series Using Recursion
- Divide and Conquer
- DSA - Divide and Conquer
- DSA - Max-Min Problem
- DSA - Strassen's Matrix Multiplication
- DSA - Karatsuba Algorithm
- Greedy Algorithms
- DSA - Greedy Algorithms
- DSA - Travelling Salesman Problem (Greedy Approach)
- DSA - Prim's Minimal Spanning Tree
- DSA - Kruskal's Minimal Spanning Tree
- DSA - Dijkstra's Shortest Path Algorithm
- DSA - Map Colouring Algorithm
- DSA - Fractional Knapsack Problem
- DSA - Job Sequencing with Deadline
- DSA - Optimal Merge Pattern Algorithm
- Dynamic Programming
- DSA - Dynamic Programming
- DSA - Matrix Chain Multiplication
- DSA - Floyd Warshall Algorithm
- DSA - 0-1 Knapsack Problem
- DSA - Longest Common Subsequence Algorithm
- DSA - Travelling Salesman Problem (Dynamic Approach)
- Approximation Algorithms
- DSA - Approximation Algorithms
- DSA - Vertex Cover Algorithm
- DSA - Set Cover Problem
- DSA - Travelling Salesman Problem (Approximation Approach)
- Randomized Algorithms
- DSA - Randomized Algorithms
- DSA - Randomized Quick Sort Algorithm
- DSA - Karger’s Minimum Cut Algorithm
- DSA - Fisher-Yates Shuffle Algorithm
- DSA Useful Resources
- DSA - Questions and Answers
- DSA - Quick Guide
- DSA - Useful Resources
- DSA - Discussion
Prim’s Minimal Spanning Tree
Prim’s minimal spanning tree algorithm is one of the efficient methods to find the minimum spanning tree of a graph. A minimum spanning tree is a sub graph that connects all the vertices present in the main graph with the least possible edges and minimum cost (sum of the weights assigned to each edge).
The algorithm, similar to any shortest path algorithm, begins from a vertex that is set as a root and walks through all the vertices in the graph by determining the least cost adjacent edges.
Prim’s Algorithm
To execute the prim’s algorithm, the inputs taken by the algorithm are the graph G {V, E}, where V is the set of vertices and E is the set of edges, and the source vertex S. A minimum spanning tree of graph G is obtained as an output.
Algorithm
Declare an array visited[] to store the visited vertices and firstly, add the arbitrary root, say S, to the visited array.
Check whether the adjacent vertices of the last visited vertex are present in the visited[] array or not.
If the vertices are not in the visited[] array, compare the cost of edges and add the least cost edge to the output spanning tree.
The adjacent unvisited vertex with the least cost edge is added into the visited[] array and the least cost edge is added to the minimum spanning tree output.
Steps 2 and 4 are repeated for all the unvisited vertices in the graph to obtain the full minimum spanning tree output for the given graph.
Calculate the cost of the minimum spanning tree obtained.
Examples
Find the minimum spanning tree using prim’s method (greedy approach) for the graph given below with S as the arbitrary root.
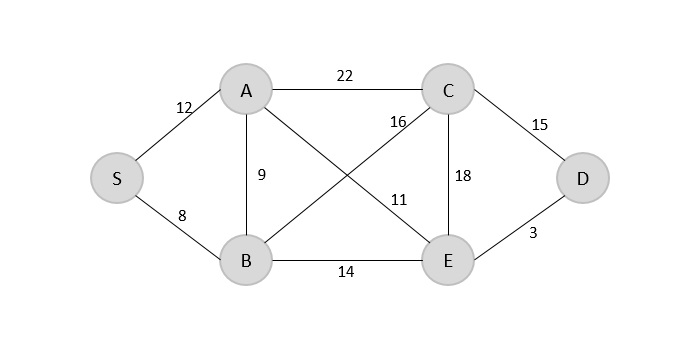
Solution
Step 1
Create a visited array to store all the visited vertices into it.
V = { }
The arbitrary root is mentioned to be S, so among all the edges that are connected to S we need to find the least cost edge.
S → B = 8
V = {S, B}
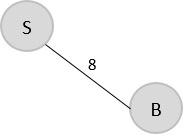
Step 2
Since B is the last visited, check for the least cost edge that is connected to the vertex B.
B → A = 9 B → C = 16 B → E = 14
Hence, B → A is the edge added to the spanning tree.
V = {S, B, A}
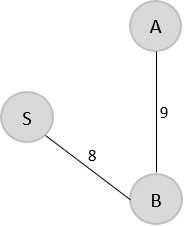
Step 3
Since A is the last visited, check for the least cost edge that is connected to the vertex A.
A → C = 22 A → B = 9 A → E = 11
But A → B is already in the spanning tree, check for the next least cost edge. Hence, A → E is added to the spanning tree.
V = {S, B, A, E}
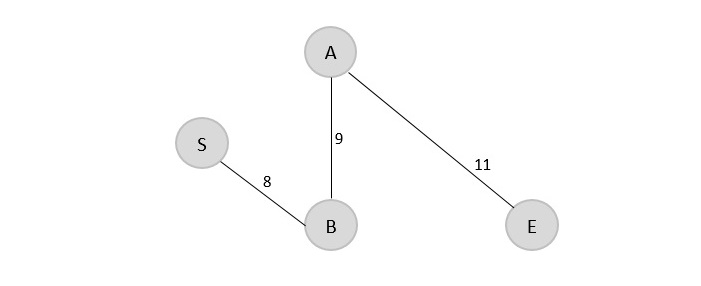
Step 4
Since E is the last visited, check for the least cost edge that is connected to the vertex E.
E → C = 18 E → D = 3
Therefore, E → D is added to the spanning tree.
V = {S, B, A, E, D}
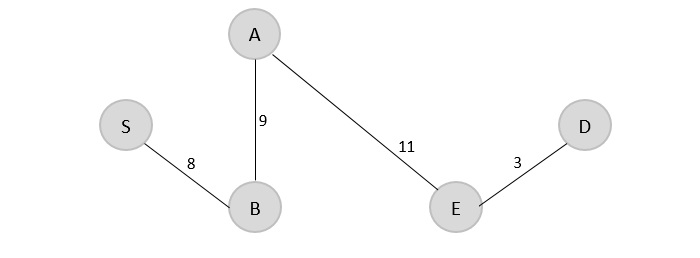
Step 5
Since D is the last visited, check for the least cost edge that is connected to the vertex D.
D → C = 15 E → D = 3
Therefore, D → C is added to the spanning tree.
V = {S, B, A, E, D, C}
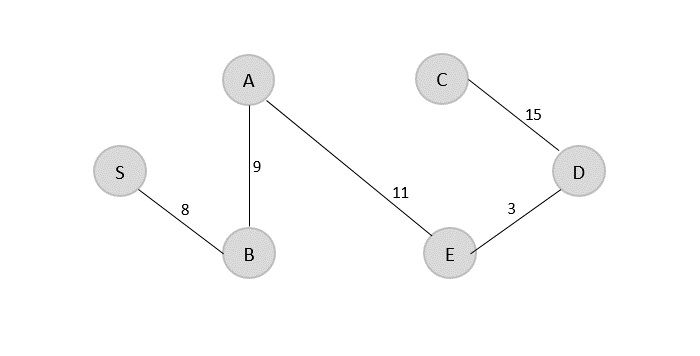
The minimum spanning tree is obtained with the minimum cost = 46
Example
The final program implements Prim’s minimum spanning tree problem that takes the cost adjacency matrix as the input and prints the spanning tree as the output along with the minimum cost.
#include<stdio.h>
#include<stdlib.h>
#define inf 99999
#define MAX 10
int G[MAX][MAX] = {
{0, 19, 8},
{21, 0, 13},
{15, 18, 0}
};
int S[MAX][MAX], n;
int prims();
int main(){
int i, j, cost;
n = 3;
cost=prims();
printf("Spanning tree:");
for(i=0; i<n; i++) {
printf("\n");
for(j=0; j<n; j++)
printf("%d\t",S[i][j]);
}
printf("\nMinimum cost = %d", cost);
return 0;
}
int prims(){
int C[MAX][MAX];
int u, v, min_dist, dist[MAX], from[MAX];
int visited[MAX],ne,i,min_cost,j;
//create cost[][] matrix,spanning[][]
for(i=0; i<n; i++)
for(j=0; j<n; j++) {
if(G[i][j]==0)
C[i][j]=inf;
else
C[i][j]=G[i][j];
S[i][j]=0;
}
//initialise visited[],distance[] and from[]
dist[0]=0;
visited[0]=1;
for(i=1; i<n; i++) {
dist[i] = C[0][i];
from[i] = 0;
visited[i] = 0;
}
min_cost = 0; //cost of spanning tree
ne = n-1; //no. of edges to be added
while(ne > 0) {
//find the vertex at minimum distance from the tree
min_dist = inf;
for(i=1; i<n; i++)
if(visited[i] == 0 && dist[i] < min_dist) {
v = i;
min_dist = dist[i];
}
u = from[v];
//insert the edge in spanning tree
S[u][v] = dist[v];
S[v][u] = dist[v];
ne--;
visited[v]=1;
//updated the distance[] array
for(i=1; i<n; i++)
if(visited[i] == 0 && C[i][v] < dist[i]) {
dist[i] = C[i][v];
from[i] = v;
}
min_cost = min_cost + C[u][v];
}
return(min_cost);
}
Output
Spanning tree: 0 0 8 0 0 13 8 13 0 Minimum cost = 26
#include<iostream>
#define inf 999999
#define MAX 10
using namespace std;
int G[MAX][MAX] = {
{0, 19, 8},
{21, 0, 13},
{15, 18, 0}
};
int S[MAX][MAX], n;
int prims();
int main(){
int i, j, cost;
n = 3;
cost=prims();
cout <<"Spanning tree:";
for(i=0; i<n; i++) {
cout << endl;
for(j=0; j<n; j++)
cout << S[i][j] << " ";
}
cout << "\nMinimum cost = " << cost;
return 0;
}
int prims(){
int C[MAX][MAX];
int u, v, min_dist, dist[MAX], from[MAX];
int visited[MAX],ne,i,min_cost,j;
//create cost matrix and spanning tree
for(i=0; i<n; i++)
for(j=0; j<n; j++) {
if(G[i][j]==0)
C[i][j]=inf;
else
C[i][j]=G[i][j];
S[i][j]=0;
}
//initialise visited[],distance[] and from[]
dist[0]=0;
visited[0]=1;
for(i=1; i<n; i++) {
dist[i] = C[0][i];
from[i] = 0;
visited[i] = 0;
}
min_cost = 0; //cost of spanning tree
ne = n-1; //no. of edges to be added
while(ne > 0) {
//find the vertex at minimum distance from the tree
min_dist = inf;
for(i=1; i<n; i++)
if(visited[i] == 0 && dist[i] < min_dist) {
v = i;
min_dist = dist[i];
}
u = from[v];
//insert the edge in spanning tree
S[u][v] = dist[v];
S[v][u] = dist[v];
ne--;
visited[v]=1;
//updated the distance[] array
for(i=1; i<n; i++)
if(visited[i] == 0 && C[i][v] < dist[i]) {
dist[i] = C[i][v];
from[i] = v;
}
min_cost = min_cost + C[u][v];
}
return(min_cost);
}
Output
Spanning tree: 0 0 8 0 0 13 8 13 0 Minimum cost = 26
public class prims {
static int inf = 999999;
static int MAX = 10;
static int G[][] = {
{0, 19, 8},
{21, 0, 13},
{15, 18, 0}
};
static int S[][] = new int[MAX][MAX];
static int n;
public static void main(String args[]) {
int i, j, cost;
n = 3;
cost=prims();
System.out.print("Spanning tree: ");
for(i=0; i<n; i++) {
System.out.println();
for(j=0; j<n; j++)
System.out.print(S[i][j] + " ");
}
System.out.println("\nMinimum cost = " + cost);
}
static int prims() {
int C[][] = new int[MAX][MAX];
int u, v = 0, min_dist;
int dist[] = new int[MAX];
int from[] = new int[MAX];
int visited[] = new int[MAX];
int ne,i,min_cost,j;
//create cost matrix and spanning tree
for(i=0; i<n; i++)
for(j=0; j<n; j++) {
if(G[i][j]==0)
C[i][j]=inf;
else
C[i][j]=G[i][j];
S[i][j]=0;
}
//initialise visited[],distance[] and from[]
dist[0]=0;
visited[0]=1;
for(i=1; i<n; i++) {
dist[i] = C[0][i];
from[i] = 0;
visited[i] = 0;
}
min_cost = 0; //cost of spanning tree
ne = n-1; //no. of edges to be added
while(ne > 0) {
//find the vertex at minimum distance from the tree
min_dist = inf;
for(i=1; i<n; i++)
if(visited[i] == 0 && dist[i] < min_dist) {
v = i;
min_dist = dist[i];
}
u = from[v];
//insert the edge in spanning tree
S[u][v] = dist[v];
S[v][u] = dist[v];
ne--;
visited[v]=1;
//updated the distance[] array
for(i=1; i<n; i++)
if(visited[i] == 0 && C[i][v] < dist[i]) {
dist[i] = C[i][v];
from[i] = v;
}
min_cost = min_cost + C[u][v];
}
return(min_cost);
}
}
Output
Spanning tree: 0 0 8 0 0 13 8 13 0 Minimum cost = 26
inf = 999999
MAX = 10
G = [
[0, 19, 8],
[21, 0, 13],
[15, 18, 0]
]
S = [[0 for _ in range(MAX)] for _ in range(MAX)]
n = 3
def main():
global n
cost = prims()
print("Spanning tree: ")
for i in range(n):
print()
for j in range(n):
print(S[i][j], end=" ")
print("\nMinimum cost =", cost)
def prims():
global n
C = [[0 for _ in range(MAX)] for _ in range(MAX)]
u, v = 0, 0
min_dist = 0
dist = [0 for _ in range(MAX)]
from_ = [0 for _ in range(MAX)]
visited = [0 for _ in range(MAX)]
ne = 0
min_cost = 0
i = 0
j = 0
for i in range(n):
for j in range(n):
if G[i][j] == 0:
C[i][j] = inf
else:
C[i][j] = G[i][j]
S[i][j] = 0
dist[0] = 0
visited[0] = 1
for i in range(1, n):
dist[i] = C[0][i]
from_[i] = 0
visited[i] = 0
min_cost = 0
ne = n - 1
while ne > 0:
min_dist = inf
for i in range(1, n):
if visited[i] == 0 and dist[i] < min_dist:
v = i
min_dist = dist[i]
u = from_[v]
S[u][v] = dist[v]
S[v][u] = dist[v]
ne -= 1
visited[v] = 1
for i in range(n):
if visited[i] == 0 and C[i][v] < dist[i]:
dist[i] = C[i][v]
from_[i] = v
min_cost += C[u][v]
return min_cost
#calling the main() method
main()
Output
Spanning tree: 0 0 8 0 0 13 8 13 0 Minimum cost = 26
